Monthly Archives: February 2012
The rich are rich because they are greedy and cheat
Abstract
Seven studies using experimental and naturalistic methods reveal that upper-class individuals behave more unethically than lower-class individuals. In studies 1 and 2, upper-class individuals were more likely to break the law while driving, relative to lower-class individuals. In follow-up laboratory studies, upper-class individuals were more likely to exhibit unethical decision-making tendencies (study 3), take valued goods from others (study 4), lie in a negotiation (study 5), cheat to increase their chances of winning a prize (study 6), and endorse unethical behavior at work (study 7) than were lower-class individuals. Mediator and moderator data demonstrated that upper-class individuals’ unethical tendencies are accounted for, in part, by their more favorable attitudes toward greed.
On and around with Ötzi’s genome
As you’re probably more than aware by now there’s a new paper on the market (yeah, 32 bucks – but worry not that I already got my hands on it) on the most loved mummy of Europe: Ötzi the Iceman.
A. Keller et al., New insights into the Tyrolean Iceman’s origin and phenotype as inferred by whole-genome sequencing. Nature 2012. Pay per view.
It looks odd indeed… but it might be explained if we assume that from that time on, secondary (post Neolithic) Bronze Age flows from the Aegean (and Central Europe) altered gradually the genetic composition of Italy. This is supported by archaeology as far as I know: even before Mycenaean Greeks, the Aegean was influencing Southern and Central Italy more and more. This trend was reinforced in the late Bronze Age (Mycenaean colonization in the South, Etruscan migration in the Center) and the Iron Age (classical Greek colonization of Magna Graecia).
All Africa autosomal analysis at Ethio Helix
I want to call your attention at the very interesting autosomal analysis of all Africa at Ethio Helix today. Surely Africa is too big and too diverse to be captured well enough in a single pan-continental analysis but there is still a lot to learn from that study.
Copper findings in Kerala reinforce the concept of Chalcolithic India
 |
| Pottery found at Ramakkalmedu |
Source: The Hindu.
Basque mtDNA
 |
| Fig. 1, showing the detailed sampling strategy |
 |
| Table 1 |
A. Gascony:
Bearn (n=56):
- H1: 11 (20%)
- H2a: 2 (4%)
- H3: 3 (5%)
- V: 3 (5%)
- HV: 2 (4%)
- U: 16 (29%)
- K: 6 (11%)
- J: 4 (7%)
- T: 2 (4%)
- X: 3 (5%)
- Singletons: H5’36, H6, H9, H59
Bigorre (n=48):
- H1: 9 (19%)
- H3: 2 (4%)
- V: 4 (8%)
- U: 11 (23%)
- K: 5 (10%)
- J: 2 (4%)
- T: 4 (8%)
- Singletons: H2a, H6, H14, H67, HV, R0, K, I, X, W, C
Chalosse (Dax district) (n=60):
- H1: 9 (15%)
- H2a: 4 (7%)
- H6: 2 (3%)
- H13: 4 (7%)
- H74: 2 (3%)
- V: 5 (8%)
- HV: 2 (3%)
- U: 13 (22%)
- K: 3 (5%)
- J: 5 (8%)
- T: 3 (5%)
- X: 3 (5%)
- Singletons: H3, H4, H5, H8, H42
B. Northern Basque Country:
Lapurdi/Baztan (Lapurtera dialectal zone) (n=58):
- H1: 15 (26%)
- H2a: 2 (3%)
- H3: 3 (5%)
- H4: 2 (3%)
- V: 8 (14%)
- U: 13 (22%)
- J: 8 (14%)
- T: 2 (3%)
- Singletons: H5, H6, H24, K, X
Lapurdi/Lower Navarre (Benafarrera dialectal zone) (n=73):
- H1: 24 (33%)
- H3: 4 (5%)
- H5: 2 (3%)
- H20: 2 (3%)
- V: 6 (8%)
- HV: 6 (8%)
- U: 13 (18%)
- K: 2 (3%)
- J: 5 (7%)
- T: 2 (3%)
- X: 4 (5%)
- Singletons: H2a, H6, H42
Zuberoa (n=61*):
- H1: 16 (26%)
- H2a: 3 (5%)
- H5: 2 (3%)
- HV: 3 (5%)
- V: 2 (3%)
- U: 14 (23%)
- K: 5 (8%)
- J: 9 (15%)
- X: 3 (5%)
- W: 2 (3%)
- Singletons: H3, T
C. Southern Basque Country South (Spanish-speaking area since 19th century):
Araba (n=56):
- H*: 2 (4%)
- H1: 18 (32%)
- H3: 6 (11%)
- V: 3 (5%)
- HV: 5 (9%)
- U: 7 (13%)
- K: 3 (5%)
- T: 5 (9%)
- J: 4 (7%)
- Singletons: H58, N1, X
Central-Western Navarre (n=64):
- H1: 10 (15%)
- H3: 12 (19%)
- H7: 2 (3%)
- V: 7 (11%)
- U: 10 (15%)
- K: 2 (3%)
- J: 3 (5%)
- T: 7 (11%)
- I: 2 (3%)
- Singletons: H*, H2a, H5, H27, H42, H49, H81, N1, X
North-Eastern Navarre (Erronkari-Salazar): (n=55)
- H*: 2 (4%)
- H1: 9 (16%)
- H3: 5 (9%)
- H42: 4 (7%)
- V: 6 (11%)
- U: 17 (31%)
- K: 2 (4%)
- T: 6 (11%)
- J: 3 (5%)
- Singleton: K
D. Southern Basque Country North (Basque-speaking area in 20th century):
Biscay (n=59):
- H1: 17 (29%)
- H2a: 6 (10%)
- H6: 2 (3%)
- H53: 3 (5%)
- V: 2 (3%)
- HV: 3 (5%)
- U: 9 (15%)
- J: 6 (10%)
- X: 2 (3%)
- Singletons: H*, H14, H17, H24, H86, T, N1, I, K
Gipuzkoa (n=57*):
- H1: 19 (33%)
- H3: 7 (12%)
- H17: 2 (4%)
- V: 3 (5%)
- U: 12 (21%)
- K: 2 (4%)
- J: 5 (9%)
- T: 2 (4%)
- X: 2 (4%)
- Singletons: H2a, H14, W
Gipuzkoa SW (n=63):
- H1: 24 (38%)
- H2a: 3 (5%)
- H3: 3 (5%)
- H6: 2 (3%)
- V: 3 (5%)
- U: 16 (25%)
- K: 2 (3%)
- J: 3 (5%)
- T: 2 (3%)
- Singletons: H4, H58, HV, X, L3’4
North-Western Navarre (n=53):
- H1:10 (19%)
- H2a: 3 (6%)
- H3: 8 (15%)
- H4: 2 (4%)
- H5: 3 (6%)
- V: 3 (6%)
- U: 12 (23%)
- T: 4 (8%)
- J: 5 (9%)
- Singletons: H24, HV, W
E. Southern Basque Country – West Biscay (Spanish speaking since old):
Enkarterriak (n=21):
- H1: 5 (23%)
- H3: 3 (14%)
- H15: 3 (14%)
- HV: 3 (14%)
- U: 2 (10%)
- Singletons: H4, H24, H87, K, X
F. Spain (areas once within the Basque ethno-cultural area):
Northern Aragon (n=29):
- H3: 5 (17%)
- H4: 2 (7%)
- HV: 3 (10%)
- U: 5 (17%)
- K: 2 (7%)
- J: 6 (21%)
- T: 3 (10%)
- Singletons: H42, V, X
Northern Burgos province (n=24):
- H1: 2 (8%)
- H3: 4 (17%)
- U: 8 (33%)
- K: 2 (8%)
- T: 2 (8%)
- J: 2 (8%)
- Singletons: H*, H4, V, L2
Cantabria (n=19):
- H1: 7 (37%)
- H3: 3 (16%)
- H5: 2 (11%)
- J: 2 (11%)
- Singletons: H27, H30, U, K, T
La Rioja (n=52):
- H*: 2 (4%)
- H1: 13 (25%)
- H3: 7 (13%)
- H5: 3 (6%)
- U: 8 (15%)
- J: 4 (8%)
- T: 5 (10%)
- K: 3 (6%)
- Singletons: H10, H13, H30, H51, H58, R0, I
Notes:
(1) * Sample size of Zuberoa is listed as 62 and Gipuzkoa as 56 but after checking and rechecking I’m pretty sure that one individual has swapped populations. So I’m assuming that n(Zuberoa)=61 and n(Gipuzkoa)=57 for all apportions.
(2) Haplogroups in italic type are not named that way (or not named at all) in PhyloTree. I am confused by this and other nomenclature of this paper and so far haven’t got time to study what they might mean. Ideas are welcomed.
(3) U means obviously U(xK). Just using the same terminology from the paper. Again, I haven’t got any time to explore how much of that U is U5b, U5a, U4 or other clades. This is in my opinion the greatest shortcoming of the paper: ignoring U almost completely.
 |
| Frequencies of mtDNA H1 |
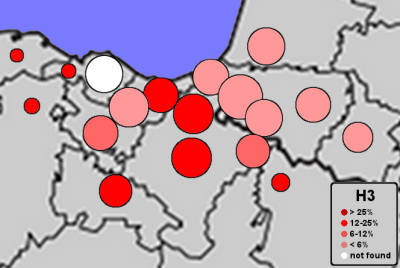 |
| Frequencies of mtDNA H3 |
 |
| Frequencies of mtDNA U(xK) |
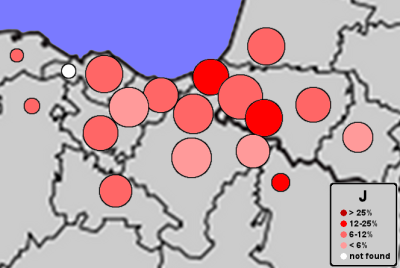 |
| Frequencies of mtDNA J |
 |
| Frequencies of mtDNA V |
Notice that V is not as common among Basques as initially reported years ago.
See also:
- Basque-specific mtDNA lineages (on U5b and J1c, argued to be also signals of post-Glacial colonization from SW Europe).
- Category ‘Basque origins’.
Oldest toy car is from Kurdistan c. 5500 BCE
Intelligence genes more elusive than Higgins’ boson
What our results show is that the way researchers have been looking for genes that may be related to intelligence — the candidate gene method — is fairly likely to result in false positives, so other methods should be used.
Maternal ancestry of Jamaicans
 |
| Jamaican slave revolt of 1759 (source) |
Late West European Neanderthals had very low matrilineal genetic diversity
Love Dalén et al., Partial genetic turnover in neandertals: continuity in the east and population replacement in the west. MBE 2012. Pay per view.
Abstract
Remarkably little is known about the population-level processes leading up to the extinction of the neandertal. To examine this, we use mtDNA sequences from 13 neandertal individuals, including a novel sequence from northern Spain, to examine neandertal demographic history. Our analyses indicate that recent western European neandertals (<48 kyr) constitute a tightly defined group with low mitochondrial genetic variation in comparison to both eastern and older (>48 kyr) European neandertals. Using control region sequences, Bayesian demographic simulations provide higher support for a model of population fragmentation followed by separate demographic trajectories in subpopulations over a null model of a single stable population. The most parsimonious explanation for these results is that of a population turnover in western Europe during early Marine Isotope Stage 3, predating the arrival of anatomically modern humans in the region.




