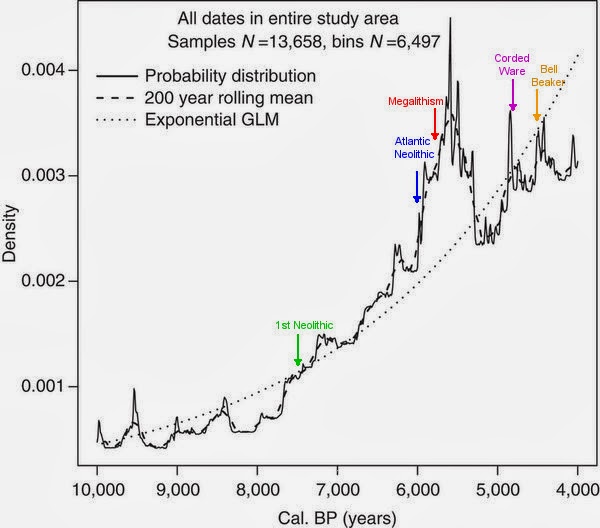
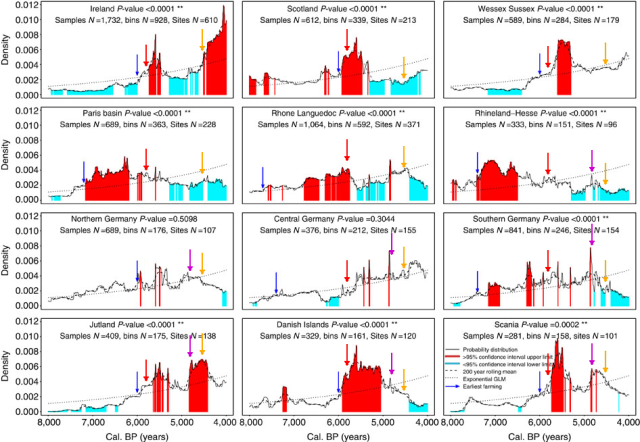
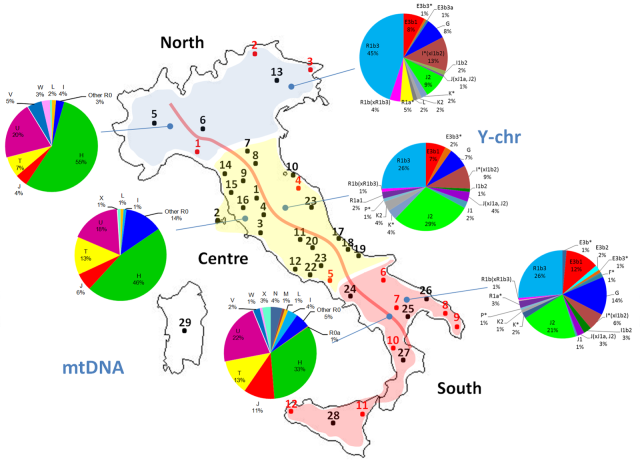
For reasons of the variegated methodology used by the various researchers this comparison across time and space has to be simplified. Still it is a valuable insight on the demographic changes produced in the Neolithic and later on in three European regions: Germany, Portugal and the Basque Country.
Germany
As you surely know already, the results of archaeogenetic sequencing in Central Europe have produced quite perplexing results: not just the Neolithic wave seems to have caused major changes but also this one was victim of similarly radical later changes in the demography. Visually:
The first period when we see an mtDNA pool similar to the modern one is already in the Late Bronze Age. However we lack data for all the early and middle Bronze Age and the data of the late Chalcolithic already points to the components of this modern pool being present, albeit in a very fragmented form. If anything there was still excess of L(xR), i.e. N(xR).
This late Chalcolithic and Bronze Age knot of Central European demographic history is still to be solved. But something we can already say for sure: the Neolithic wave was of dramatic consequences in this region but itself was wiped out by later, still ill understood, secondary waves.
Portugal
This area is not so well documented, however the data we do have still provide a very interesting sequence of the demographic history of SW or West Iberia. Visually again:
One of the problems here is, quite evidently, that sequencing only the HVS-I region is not at all enough to identify some very important haplogroups, particularly H. We can reasonably think however that most or even all of the R* sequences are actually H.
We see some but not-so-radical changes with the arrival of Neolithic: some apparent decrease of U (halved) and L(xR), a +33% growth of H and first detection of HV0 (probably V). However these changes seem to have been partly countered by Chalcolithic, plausibly by means of blending between first farmers and more purely aboriginal populations. Overall I am very much tempted to think that the arrival of Neolithic to (South and Central) Portugal only caused mild demic changes.
This fact, together with the extremely high frequencies of haplogroup H and the key role played by SW Iberia in the formation of Dolmenic Megalithism, as well as their pivotal role in Bell Beaker, including the existence of a major civilization (Zambujal, VNSP), the first one ever in Atlantic Europe, makes this area highly suspect as a possible origin for the spread of mtDNA H in Western Europe to the frequencies that we find today (c. 40-50%).
However we have only very limited archaeogenetic data from other Atlantic Megalithic regions and in general from Megalithic burials and it is at least possible that Armorica (Brittany, West France) or Denmark and the nearby Low Germany regions played important roles in this spread, which we see so dramatically exemplified in the German Bell Beaker sample.
When the finger points to the Moon, the fool looks at the finger. Portugal could be the Moon but it may just be the finger, so I will remain cautious at this stage of research. Whatever the case it does seem to me that Megalithism is a likely source of that excess H (Bell Beaker being just the finger here, almost for sure).
I must add that there seem to be some important demic changes since Chalcolithic in Portugal. Tentatively I will attribute them to the intrusive SW Iberian “horizons” (proto-Tartessian?) and/or the Luso-Celtic invasions of the Iron Age.
Basque Country
My main aim in all this compilation was, as in a sense in all my diving into prehistoric research for so many years now, to find an answer to the mystery of the origin of Basques and Basque language.
In the last few years we have been blessed with some important and revealing archaeogenetic research in this area, and therefore I could build also an informative graph for the Basque Country:
Very synthetically, I think that we can see here, much as in Portugal, some not too radical changes with the Neolithic arrival, and then relative stability until present day. This is coherent with the Basque Country not having suffered effective Indoeuropean invasions, unlike Portugal.
However I strongly feel the need to look at the fine detail in the Basque Neolithic transition, because it has some interesting question marks:
Seen as that, it would seem like the Neolithic-induced demic change was more important in Navarre and less in the Western Basque Country. However the two Ebro basin sequences (both Fuente Hoz and Los Cascajos) are very high in U* and low in U5, which is so far the only U subclade sequenced in the Paleolithic of the Basque-Cantabrian area. At this point I do not really know how to interpret this fact nor even what kind of U sublineage is that one.
What I do know is that, on one side, the Biscay-Gipuzkoan area seems to have been initially unaffected by Neolithic demic waves and that the Paternabidea sequence is very very similar to modern day Basque average (and even more in its own sub-region).
It is very possible that the Basque periphery, notably the Ebro banks, suffered more intense demic changes than the core Basque areas of the piedmont. However, when compared with other European regions (very especially Central Europe) the Basque genetic pool seems quite stable since Neolithic times.
Is Basque language Neolithic?
Even if genetics and language need not to be tightly related, of course, the question of the origin of Basque language and the proposed Vasconic language family, believed to have been spoken in much of Europe at some point in Prehistory, are indeed related to the genetic origin of the Basque people.
There are four main models for the origin of Basque and Vasconic:
- Magdalenian (Paleolithic) origin in the Franco-Cantabrian region some 17-15,000 years ago (incl. possible sub-waves like Tardenoisian/geometric Epipaleolithic).
- Neolithic origin.
- Megalithic origin.
- More or less recent (Iron Age?) arrival, defended by mostly by the fanatics of Indoeuropean continuity.
We can safely discard #4 only based on archaeology but the genetic aspect seems to add even more weight to this dismissal, after all it is Indoeuropean speaking peoples the ones which show obvious signs of demic change, sometimes very dramatic, not Basques.
Personally, and with due caution, I would also cast doubt on #1, partly because the Vasconic substrate area seems to include strongly many parts of Italy
like Sardinia, in principle unaffected by the Magdalenian expansion, and I would also include at least to some extent parts of the Balcans (for example the Ibar river in Kosovo).
So I am rather inclined for model #2, i.e. that Vasconic was the language family spoken by European Neolithic peoples with roots in Thessaly (pre-Sesklo→Mediterranean Neolithic, proto-Sesklo→Balcano-Danubian Neolithic). I cannot of course exclude a possible re-expansion of some of those languages within the Atlantic Megalithic phenomenon, which I would deem responsible of the expansion of much of mtDNA H up to modern frequencies, however I doubt this one is the source because it is difficult to explain the presence of Vasconic in many pockets in which Megalithism was at best very secondary or did not exist at all (for example most of the Ancient Iberian area, Sardinia, the Balcans, etc.)
So my tentative proposal is that there was a root Vasconic spoken some 9000 years ago in Thessaly (Northern Greece), which split (as per archaeology) in two branches:
- Southern or Western Vasconic (Impressed-Cardium Pottery and related cultures, including the Megalithic urheimat in Portugal).
- Northern or Eastern Vasconic (Red-White Painted Ware in the Balcans and later Linear Pottery in Central Europe).
Basque, ancient Sardinian, Iberian and the hypothetical lingua franca associated to Megalithism would belong to Southern Vasconic. Danubian Neolithic peoples would have spoken Northern Vasconic instead but, as we can see, they were eventually all but wiped out by secondary arrivals from West and East. Even the very Balcanic core areas of Thessaly, Macedonia and Serbia also suffered an invasion early on by peoples with Beige-Black pottery (Vinca-Dimini) surely related to Tell Halaf. So the main survivor to the Metal Ages was Southern (Western) Vasconic, which was then wiped out (excepted Basque) by the Indoeuropean invasions of Celtic and Italic peoples.
We can still see fossils however. One of my favorite examples is the Latin particle bi- (as in bilateral, bilingual, etc.), which seems derived from Vasconic bi (two, at least in modern Basque) and unrelated to PIE *dwos. Also the English words kill and ill, which seem related to Basque verb hil(-du) (pronounced /hill/ or /ill/ and meaning to die or to kill, depending on how you conjugate it). Again both English terms do not have any apparent PIE origins, although they may derive from proto-Germanic. These are just examples, of course, there seems to be much more to be researched.
Appendix: detail of the data and bibliography:
LINK.