 |
| Fig 2 (legend) |
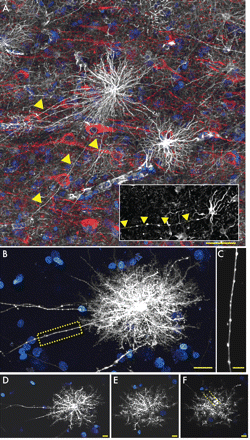
One of the most striking features distinguishing humans and chimpanzee from other lower primate and rodent astrocytes was the presence of a previously undescribed pool of morphologically distinct GFAP+ cells residing in layers 5–6, characterized by long fibers with prominent varicosities (Fig. 2A). (…) In our analysis of primate tissue, we were able to locate a small number of varicose projection astrocytes within layers 5 or 6 of the chimpanzee cortex (Fig. 2A, inset). These cells differed from those seen in human in that they were smaller and less complex, with fewer main GFAP+ processes.
In addition to being more numerous than their chimpanzee counterparts, the morphology of interlaminar astrocytes is subtly different in humans. Human interlaminar astrocytes have small spheroid cell bodies and several short processes that contribute to the pial glial limitains, creating a thick network of GFAP fibers not seen in the primate.
… the average diameter of protoplasmic cortical astrocytes in the chimpanzee brain was 81.7 ± 1.9 μm (n = 36), which is significantly smaller than human astrocytes, but significantly larger than protoplasmic astrocytes in mouse brain…
Glial cells outnumber neurons by at least 6 to 1 but the ratio differs
in different parts of the nervous system. The ratio can be 100 glials to
1 neuron along nerves in the white matter tracts in the brain; in the
frontal cortex the ratio is 4 to 1. Interestingly, whales and dolphins
have 7 glials for every neuron in their gigantic forebrains. (Fields, R.
Douglas, PhD. The Other Brain. P P 24. NY:Simon & Schuster, 2009.)








